All RIS elements start with a function and, therefore, must be chosen among the sequences of the heads. Thus, a point is randomly chosen in the head and, from this point onwards, the gene is scanned until a function is found. This function becomes the start position of the RIS element. If no functions are found, the operator does nothing. The transposition operator randomly chooses the chromosome, the gene to be modified, the start of the RIS element, and its length. Typically, a root transposition rate
pris of 0.1 and a set of three RIS elements of different sizes are used.
In the example shown in Figure 3.16, a pris = 1.0 was used together with three RIS elements of lengths 2, 3, and 5. In order to simplify the analysis of the effects of RIS transposition, all the other search operators were switched off. By the results shown in
Figure 3.16 you can see that this operator alone is also capable of making populations evolve. In this case, by generation 17 a perfect solution to the Majority
(a, b, c) function problem was discovered (chromosome 8). This individual is a descendant of chromosome 5 of generation 16. In this case, the RIS element “AcAaa” in gene 1 (positions 7-11) was transposed to the start position of that gene
(Figure 3.17). As in IS transposition, in RIS transposition a sequence with exactly the same length as the transposon is deleted at the end of the head in order to maintain the structural organization of the chromosome. In this case, the sequence “AaAcA” was deleted.
|
Generation N: 0
012345678901234567890012345678901234567890
NAccbAaOccaaaaabccccbOAAaANObAbababcacccab-[0] = 4
NOONAONbAAcbbaabbbbccAOONaOacAOaccbaacacac-[1] = 4
AbabNcOcacaaaccbbbcccNcAbOaObcOabbbccbbcaa-[2] = 3
NNAcaNNANbccbcbbbacbcOacAabaNccbbbacbcbbcb-[3] = 6
ONbcaNNcAAccbcbbcaabcAaOaabbbAbaabccabbbca-[4] = 5
NObcNNaaNbabccccaccacNbcAbcNaAbacaaabcabab-[5] = 2
NOAacOcbbAacbaacaaabcAOacNaaabOcacccabbcbb-[6] = 4
ONaAcaNcOacbbbacbccacOcOAOcNcANcbcaabbbaba-[7] = 4
AOAONbAObbbbcbbacbbcbOONAbANAcbbcbabcccaab-[8] = 4
ANbbcbaaccaaaaccbabcbAOaaacObaObabccaacaba-[9] = 6
...
Generation N: 16
012345678901234567890012345678901234567890
AOcAcAOccNaaaaabccccbAbaabAbaOAababcacccab-[0] = 7
AaAcAaOAaOaaaaabccccbAOAbaObAObababcacccab-[1] = 7
OcOccAaOccaaaaabccccbAaAAbaabAaababcacccab-[2] = 7
AaOccAcAaOaaaaabccccbAOObAAOAObababcacccab-[3] = 7
OAaaaAaAaAaaaaabccccbObAObAAbaOababcacccab-[4] = 6
OAaOaAaAcAaaaaabccccbAAbObAObAAababcacccab-[5] = 7
AaAaAcAaOAaaaaabccccbObAObAAbaOababcacccab-[6] = 7
OccAcAaOccaaaaabccccbObAAOAObAAababcacccab-[7] = 6
OccAaOccAcaaaaabccccbObAAOAObAAababcacccab-[8] = 6
OccOccAaOcaaaaabccccbAaAAbaabAaababcacccab-[9] = 7
Generation N: 17
012345678901234567890012345678901234567890
OccOccAaOcaaaaabccccbAaAAbaabAaababcacccab-[0] = 7
AaOAaAaAcAaaaaabccccbObAObAAbaOababcacccab-[1] = 6
OAAaAaAcAaaaaaabccccbObAObAAbaOababcacccab-[2] = 6
AaOcOccAaOaaaaabccccbAaAAbaabAaababcacccab-[3] = 7
OaAaAOAaOaaaaaabccccbAAbObAObAAababcacccab-[4] = 7
OccAaOccOcaaaaabccccbAaAAbaabAaababcacccab-[5] = 7
AaAaAcAaOAaaaaabccccbAbObAObAAbababcacccab-[6] = 7
OcOccAaOccaaaaabccccbAAAaAAbaabababcacccab-[7] = 7
AcAaaOAaOaaaaaabccccbAAbObAObAAababcacccab-[8] = 8
AaOccAcAaOaaaaabccccbAAOAOObAAOababcacccab-[9] = 6
|
Figure 3.16. An initial population and its later descendants created via RIS transposition to solve the Majority
(a, b, c) function problem. The chromosomes encode sub-ETs linked by OR. Note that none of the later descendants resemble their ancestors of generation 0. Note also the appearance of repetitive sequences in later generations. The perfect solution found by generation 17 (chromosome 8) and its mother (chromosome 5 of generation 16) are shown in
blue. The event of transposition that led to this perfect solution can be seen in
Figure 3.17.
It is worth emphasizing that this highly disruptive operator is, nevertheless, capable of forming simple, repetitive sequences like, for instance, the sequences (Aa)n or (Oc)n present in the later generations of
Figure 3.16. Interestingly, DNA is also full of repetitive sequences. In fact, in some eukaryotes more than 40% of DNA consists of small repetitive sequences. Most of these sequences are not even transcribed, but some genes are also interspersed with small islands of repetitive sequences.
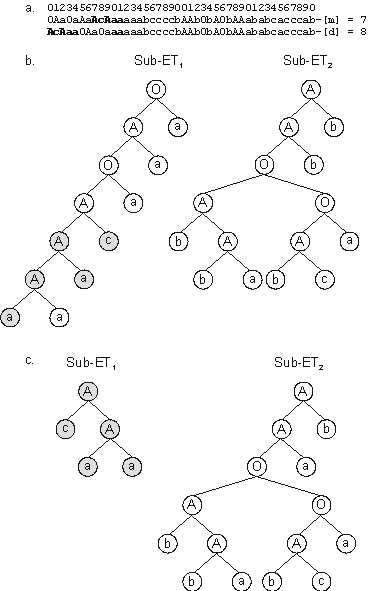
Figure 3.17. Illustration of RIS transposition and its effects.
a) An event of RIS transposition with the transposon shown in bold. Note that a sequence with the length of the transposon is deleted at the end of the head of the target gene. Note also that the transposon became, in this case, only partially duplicated in the daughter chromosome; its other elements were deleted.
b) The sub-ETs encoded by the mother chromosome (before RIS transposition).
c) The sub-ETs encoded by the daughter chromosome (after RIS transposition). The transposon elements are shown in gray. Note that, in this case, root transposition changed drastically the
sub-ET1, shortening the mother sub-ET in eight nodes.
|
