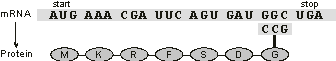|
The logistics of the synthesis of both DNA and RNA, based on the complementarity of nucleotides, is fairly simple when compared with the synthesis of proteins. Chemically, there is no simple way of pairing directly the triplet codons with the appropriate amino acids. Indeed, most of the sophisticated machinery of translation evolved to solve exactly this problem. The amino acid must be correctly attached to special molecules which are then coupled to the correct codons in the mRNA molecule. This special class of molecules are also RNA molecules (transfer RNAs), but they are structurally and functionally very different from mRNA. As we have seen, tRNAs have tertiary structure and can therefore have varied functionalities. Although the identification of the correct amino acids is not made by the tRNAs themselves, their unique three-dimensional structure is fundamental to their correct identification by particular enzymes. Each such enzyme recognizes both a particular amino acid and the appropriate tRNA, further attaching the amino acid to the tRNA. Furthermore, each tRNA contains also a nucleotide sequence (the anticodon) that is complementary to the appropriate codon, through which the coupling of the correct amino acid carrier to the mRNA is made.
The set of rules to translate the triplet codons of mRNA into amino acids is the genetic code.
Figure 1.6 shows the 64 codons and the amino acids or instructions each codes for. The amino acids are represented by a three-letter abbreviation and by a one-letter abbreviation often used in describing the primary structure in proteins. (Iíll stick to this one-letter abbreviation to represent protein chains in all the figures of this chapter.)
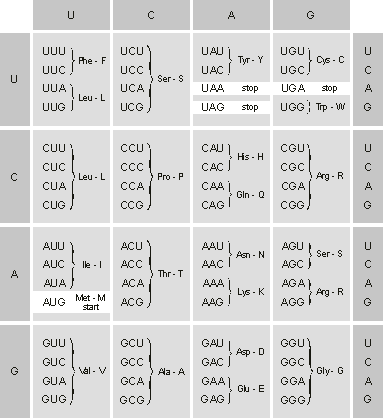
Figure 1.6. The genetic code as expressed in mRNA. Three of the 64 codons are stop signals. The start codon also codes for metionine. Note that the code is redundant with many codons coding for the same amino acid. The amino acid corresponding to each codon is given by the three-letter and one-letter abbreviation often used in describing amino acid sequences in proteins.
The message in the mRNA molecule is read one codon at a time, each codon being correctly paired with the appropriate tRNA through the anticodon, and the transported amino acids are linked one after another, forming a long, linear protein chain whose sequence exactly reflects the sequence of the gene
(Figure 1.7). It is worth mentioning that ribosomes Ė key particles in this complex process of protein synthesis Ė are huge macromolecular structures composed of numerous proteins and another class of RNA molecules, ribosomal RNAs. Like tRNAs, rRNAs also have unique three-dimensional structures. Not surprisingly, rRNAs also participate as real enzymes in the myriad of chemical reactions that occur in the ribosomal machine.
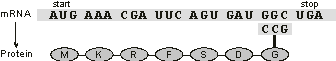
Figure 1.7. An oversimplification of translation, showing the essential elements: the mRNA template, the start and stop codons, a charged tRNA, and the emergent protein chain. The sequence shown here is unrealistically short to display both the start and stop codons.
Fortunately for us, the chemical intricacies of translation are of limited interest in a computer system like GEP. In a computer system as such, the rules of translation (in the broader sense) are simply defined and simply applied: we donít have to deal with chemistry. Indeed, we donít have to deal with either intermediate transcription processes or with complicated genetic codes requiring complicated translation mechanisms. The genetic code of GEP is a simple one-to-one relationship between the symbols of the genome and the functions and variables (also called terminals or leaves in the jargon of evolutionary computation) they represent.
|