Homeotic genes have exactly the same kind of
structure as conventional genes and are built using an identical
process. They also contain a head and a tail domain, with the heads
containing, in this case, linking functions (so called because they
are actually used to link different ADFs) and a special class of
terminals – genic terminals – representing conventional genes,
which, in the cellular system, encode different ADFs; the tails
contain obviously only genic terminals.
Consider, for instance, the following chromosome:
|
01234567801234567801234567801234567890 |
|
|
/-b/abbaa*a-/abbab-*+abbbaa**Q2+010102 |
(11) |
It codes for three conventional genes and one homeotic gene
(shown in blue). The conventional genes encode, as usual, three
different sub-ETs, with the difference that now these sub-ETs will
act as ADFs and, therefore, may be invoked multiple times from
different places. And the homeotic gene controls the interactions
between the different ADFs (Figure 8). As you
can see in Figure 8, in this particular case,
ADF0 is used twice in the main program, whereas ADF1
and ADF2 are both used just once.
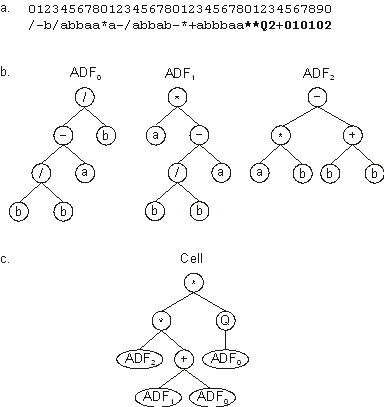
Figure 8. Expression of a unicellular system with three
Automatically Defined Functions. a) The chromosome composed
of three conventional genes and one homeotic gene (shown in bold).
b) The ADFs codified by each conventional gene. c) The
main program or cell.
It is worth pointing out that homeotic genes have their specific
length and their specific set of functions. And these functions can
take any number of arguments (functions with 1, 2, 3, …, n,
arguments). For instance, in the particular case of
chromosome (11), the head length of the homeotic gene hH
is equal to five, whereas for the conventional genes h = 4;
the function set used in the homeotic gene FH consists of
FH = {+, -, *, /, Q}, whereas for the conventional genes
the function set consists of F = {+, -, *, /}. As shown in
Figure 8, this cellular system is not only a
form of elegantly allowing the evolution of linking functions in
multigenic systems but also an extremely elegant way of encoding
ADFs that can be called an arbitrary number of times from an
arbitrary number of different places.
|
