The structural organization of GEP genes is better understood in terms of open reading frames (ORFs). In biology, an ORF or coding sequence of a gene begins with the start codon, continues with the amino acid codons, and ends at a termination codon. However, a gene is more than the respective ORF, with sequences upstream of the start codon and sequences downstream of the stop codon. Although in GEP the start site is always the first position of a gene, the termination point does not always coincide with the last position of a gene. Consequently, it is common for GEP genes to have non-coding regions downstream of the termination point. (For now we will not consider these non-coding regions, as they do not interfere with expression.)
Consider, for example, the algebraic expression:
|
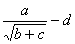
|
(1) |
It can also be represented as a diagram or ET:
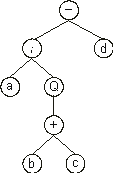
where “Q” represents the square root function.
This kind of diagram representation is in fact the phenotype of GEP chromosomes. And the genotype can be easily inferred from the phenotype as follows:
which is the straightforward reading of the ET from left to right and from top to bottom (exactly as we read a page of text). The
expression (2) is an open reading frame, starting at “-” (position 0) and terminating at “c” (position 7). These ORFs were named
K-expressions from Karva language.
Consider another open reading frame, the following K-expression:
|
012345678901 |
|
|
/Q**a*+baaba |
(3) |
Its expression as an ET is also very simple and straightforward. In order to express the ORF correctly, we must follow the rules governing the spatial distribution of functions and terminals. First, the start of a gene corresponds to the root of the expression tree which is placed in the topmost line. Second, in the next line, below each function, are placed as many branch nodes as there are arguments to that function. Third, from left to right, the nodes are filled consecutively with the next elements of the K-expression. Fourth, the process is repeated until a line containing only terminals is formed. In this case, the following expression tree is formed:
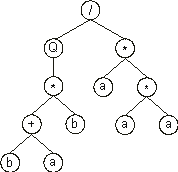
which mathematically corresponds to 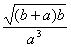 . .
Looking at the structure of GEP ORFs only, it is difficult or even impossible to see the advantages of such a representation, except perhaps for its simplicity and elegance. However, when open reading frames are analyzed in the context of a gene, the advantages of this representation become obvious. As previously stated, GEP chromosomes have fixed length, and they are composed of one or more genes of equal length. Consequently, the length of a gene is also fixed. Thus, in gene expression programming, what varies is not the length of genes which is constant, but the length of the ORF. Indeed, the length of an open reading frame may be equal to or less than the length of the gene. In the first case, the termination point coincides with the end of the gene, and in the latter, the termination point is somewhere upstream of the end of the gene.
What is the function of these non-coding regions of GEP genes? We will see that they are the essence of gene expression programming and evolvability, for they allow the modification of the genome using several genetic operators without restrictions, always producing syntactically correct programs. Thus, in gene expression programming, the fundamental property of genotype/phenotype systems – syntactic closure – is intrinsic, allowing the totally unconstrained restructuring of the genotype and, consequently, an efficient evolution.
In the next section we are going to analyze the structural organization of GEP genes in order to understand how they invariably code for syntactically correct programs and why they allow an unconstrained application of virtually any genetic operator.
|

